GCAT CATALOGUE DATA
The shared use of the GCAT data is one of the objectives of the GCAT project. For this reason, you can find the GCAT project on the Maelstrom Catalogue. Maelstrom Research's activities are founded on the data harmonization methodology developed under the DataSHaPER project, the open-source software developed by the OBiBa team, and the federated data analysis methodology developed under the DataSHIELD project. See our project on the Maelstrom Catalogue here.
Moreover, the GCAT data variables definition, as well as summary data, can be browsed on the GCAT variable catalogue on MICA to help identify variables of interest to support your collaborative research projects. A short video tutorial showing how to use the variable browser is available below (use CC button for video captions).
GCAT variable search from GCAT Genomes for Life on Vimeo
Avalaible GCAT data
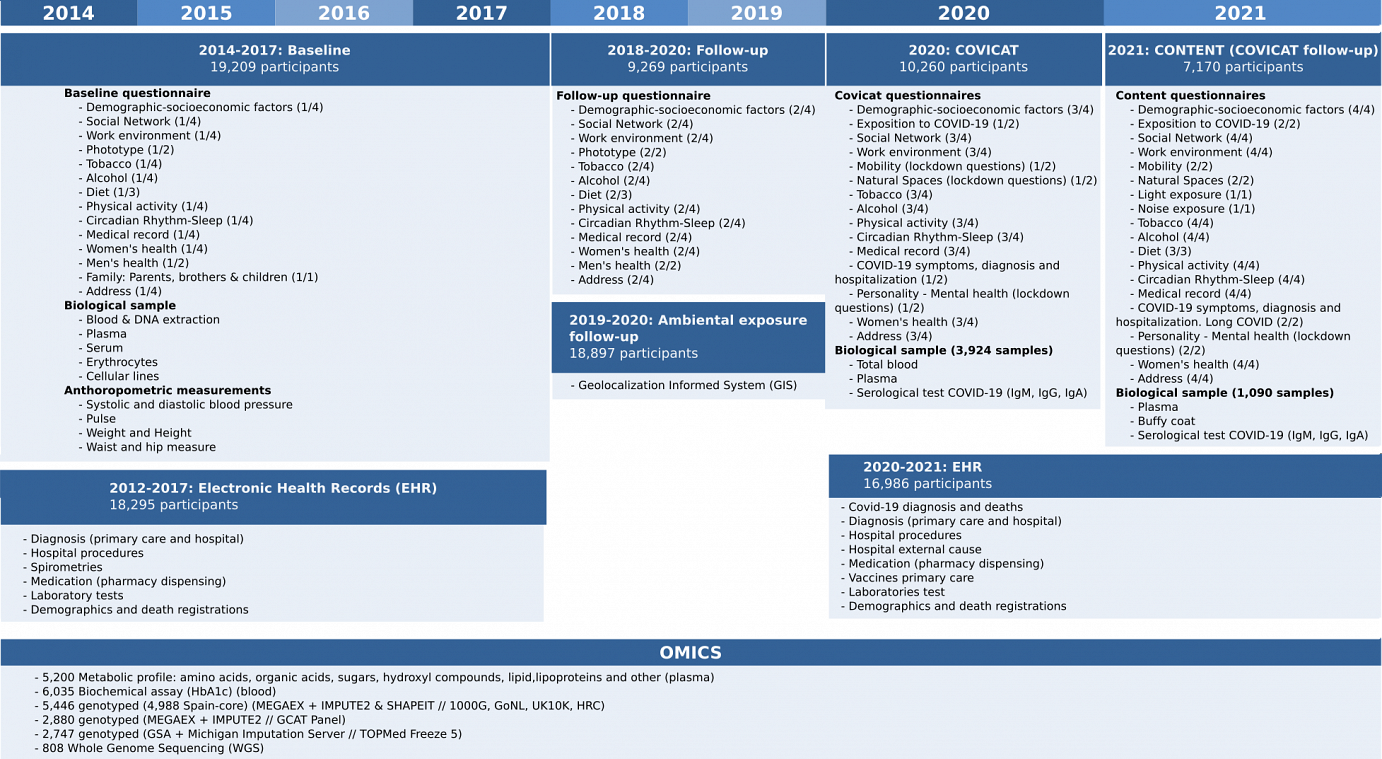
For more details, here you can find an interactive summary table of all available GCAT data and a summary table with Baseline Questionnaire and Follow up Variables.
GCAT health data
For health data avalaible on the GCAT, we have used the ICD-9 disease classification. Here you can find interactive tables summarizing autoreported diseases and family history diseases at Baseline.
Please download the main descriptive GCAT paper for more information. Cohort Profile: GCAT Genomes for Life. A Prospective Cohort study of the Genomes of Catalonia. Obón-Santacana M, Vilardell M, Carreras A, Durán X, Velasco J,Galván-Femenía I, Alonso T, Puig L, Sumoy L, Duell, E.J, Perucho M, Moreno V, de Cid R (link)
Summary of all available GCAT data
|
Data Type |
Number of participants |
Details |
Date of adquisition |
|
Baseline assessment |
Whole cohort |
Questionnaires, Physical measures, samples |
2014-2017 |
|
Repeat of baseline assessment |
Whole cohort |
Questionnaire follow-up every 2 years |
2018 |
|
Genotyping (baseline samples) |
5.459 (GCATcore) |
Dense genotyping array with 666.695 markers after quality control |
2016 |
|
Genotyping extended (baseline samples) |
5.459 (GCATcore) |
Dense genotyping map with 15,078.461 variants by in silico imputation (IMPUTE) |
2017-2018 |
|
Food frequency web questionnaire (follow-up) |
Whole cohort |
Participants are invited by email to provide additional information about diet; estimates of nutrient intake |
2017-2018 |
|
Biochemical assay (baseline |
6.000 |
Glycated haemoglobin (haemoglobin |
2016-2017 |
|
Metabolome (baseline |
5.000 (GCATcore) |
Biomarkers with known disease |
2017-2018 |
|
Chronotype web |
Whole cohort |
Participants are invited by email to provide additional information (ie, sleep behavior, circadian rhythm, and work shift) |
2017-2018 |
|
Exposome (baseline) |
Whole cohort |
Map of environmental exposures |
2017-2018 |
|
Other web-based |
Whole cohort |
Participants are invited by email to provide additional information via web about working places. Information |
2017-2018 |
|
Exome |
200 (GCATcore) |
Clinic custom exome of hereditary cancer in 126 hereditary cancer genes (400×) |
2017 |
|
Whole-genome sequencing |
808 |
30× whole genome sequencing from 1000 volunteers, 20% from GCATcore |
2017-2018 |
|
Epigenome |
150 |
DNA methylation epigenomic profile using Infinum Methylation EPIC 850K beadarray assay |
2018 |
|
Health record linkage |
|||
|
Primary care |
Whole cohort |
ICD/ATC/ OPCS procedures/laboratory |
2017-2021 |
|
Death registrations |
Whole cohort |
ICD-coded cause specific mortality |
2017-2021 |
|
Hospital inpatient |
Whole cohort |
ICD/ATC/OPCS procedures/laboratory |
2017-2021 |
|
Hospital outpatient |
Whole cohort |
ICD (few)/OPCS |
2018-2021 |
| COVID related information | Whole cohort | COVID tests, diagnoses, hospitalizations, vaccination | 2020-2021 |
|
Other |
Whole cohort |
National mental healthcare/ |
2018 |
|
Whole cohort |




